Playing video gamesisn't exactly rocket science but, thanks toFoldit, it can be molecular biology.
排序的。
Advertisement
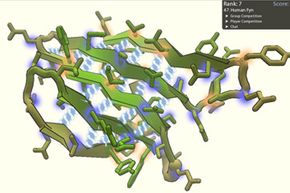
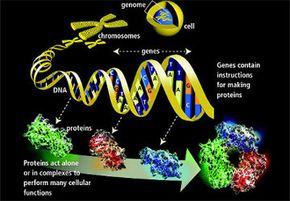
Developed in 2008 by University of Washington researchers, the multiplayer online game increases the human mind's knack for spatial reasoning by the brainpower of tens of thousands of players. Solo and in teams, these amateur analysts vie to crack the most perplexing puzzles vexing molecular biologists today: how individual proteins and their component amino acids fold.
The answer is worth more than bragging rights or achievement unlocks (gaming speak for when a player completes a particularly awesome task); it provides the means to deliver medicines that could one day stop diseases like human immunodeficiencyvirus(HIV) dead in their tracks.
It's no wonder such a buzz ensued when a 2011 publication in the journal Nature Structural & Molecular Biology reported that Foldit players had unraveled a key protein inMason-Pfizer monkey virus(MPMV), the simian version of HIV, that had stymied researchers for more than a decade.
Like John Henry versus the steam hammer or Garry Kasparov versus Deep Blue, Foldit players showed that humans still have a thing or two to teach machines; unlike Henry, who died, or Kasparov, who lost in a rematch, the protein-folding gamers still have an edge over the brute-force number crunching of supercomputers.
To understand the scope of this achievement and what it could mean for the future of HIV, let's look at why understanding how a protein folds is so important.
Advertisement